In vivo imaging of immediate early gene expression dynamics segregates neuronal ensemble of memories of dual events
Abstract
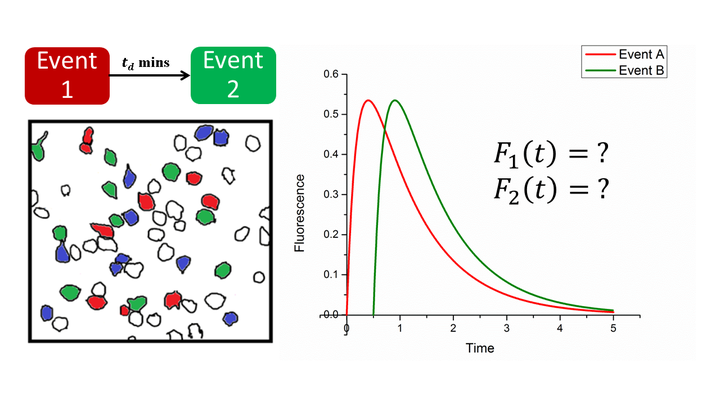
Identification of neurons undergoing plasticity in response to external stimuli is one of the pertinent problems in neuroscience. Immediate early genes (IEGs) are widely used as a marker for neuronal plasticity. Here, we model the dynamics of IEG expression as a consecutive, irreversible first-order reaction with a limiting substrate. First, we develop an analytical framework to show that such a model, together with two-photon in vivo imaging of IEG expression, can be used to identify distinct neuronal subsets representing multiple memories. Using the above combination, we show that the expression kinetics, rather than intensity threshold, can be used to identify neuronal ensembles responding to the presentation of two events in vivo. The analytical expression allowed us to segregate the neurons based on their temporal response to one specific behavioural event, thereby improving the ability to detect plasticity related neurons. We image the retrosplenial cortex (RSc) of cfos-GFP transgenic mice to follow the dynamics of cellular changes resulting from contextual fear conditioning behaviour, enabling us to establish a representation of context in RSc at the cellular scale following memory acquisition. Thus, we obtain a general method that distinguishes neurons that took part in multiple temporally separated events by measuring fluorescence of individual neurons in live mice.